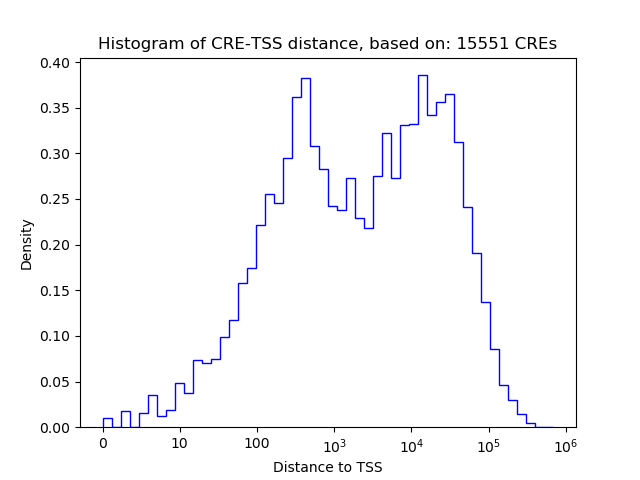
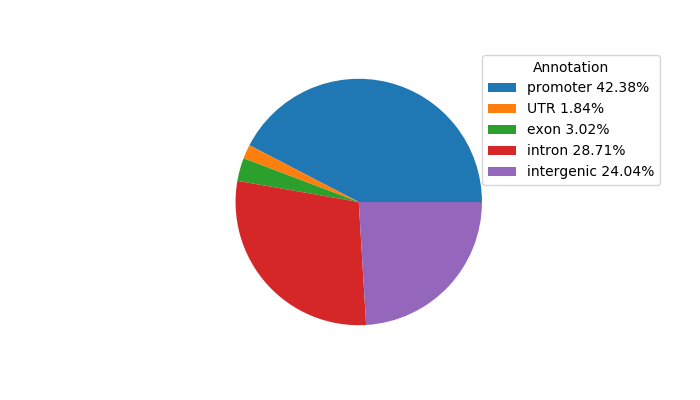
Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
Results for Glis2
Z-value: 4.69
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Glis2 | chr16_4586338_4587480 | 7804 | 0.126187 | -0.82 | 1.5e-14 | Click! |
| Glis2 | chr16_4596088_4596431 | 1546 | 0.276938 | 0.66 | 5.5e-08 | Click! |
| Glis2 | chr16_4594874_4595091 | 269 | 0.866560 | 0.59 | 2.5e-06 | Click! |
| Glis2 | chr16_4595091_4596084 | 874 | 0.473690 | 0.56 | 7.5e-06 | Click! |
| Glis2 | chr16_4598925_4599076 | 2163 | 0.205795 | 0.54 | 2.4e-05 | Click! |
Showing 1 to 5 of 5 entries
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_124439906_124440949 | 60.65 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
441 |
0.79 |
| chr12_3236518_3237725 | 43.13 |
Rab10os |
RAB10, member RAS oncogene family, opposite strand |
510 |
0.74 |
| chr14_3948585_3949537 | 23.40 |
Gm3095 |
predicted gene 3095 |
14486 |
0.11 |
| chr14_4649279_4649596 | 19.89 |
Gm3239 |
predicted gene 3239 |
14755 |
0.11 |
| chr14_5000842_5001422 | 19.52 |
Gm3298 |
predicted gene 3298 |
14759 |
0.13 |
| chr14_7488846_7489218 | 18.65 |
Gm3752 |
predicted gene 3752 |
5270 |
0.15 |
| chr14_6037512_6038662 | 18.23 |
Gm8206 |
predicted gene 8206 |
122 |
0.93 |
| chr14_4110111_4111235 | 18.19 |
Gm8108 |
predicted gene 8108 |
147 |
0.94 |
| chr2_107125684_107125835 | 17.95 |
Gm13903 |
predicted gene 13903 |
1799 |
0.38 |
| chr10_26078964_26079158 | 17.55 |
Tmem200a |
transmembrane protein 200A |
9 |
0.95 |
Network of associatons between targets according to the STRING database.
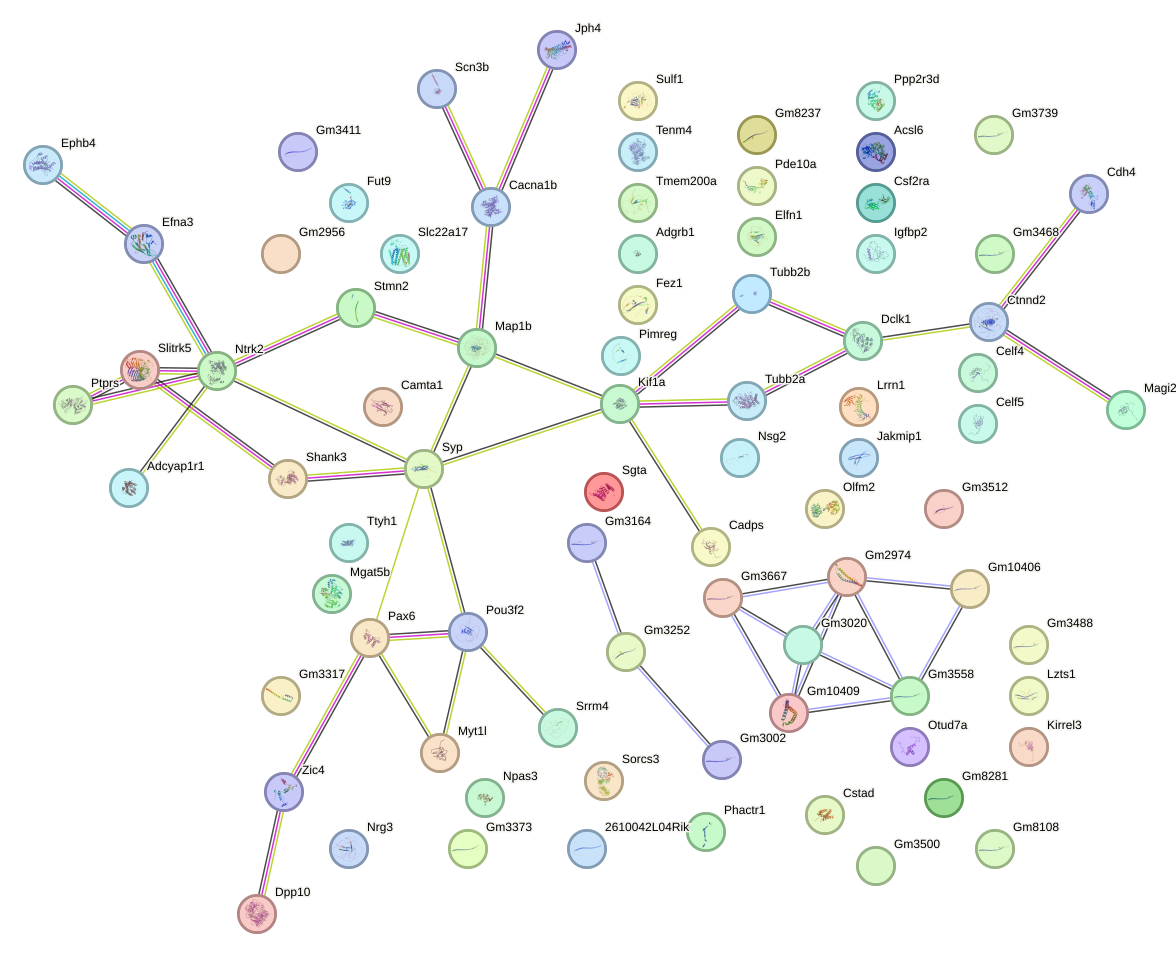
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 69.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.9 | 62.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 2.9 | 32.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 1.6 | 32.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 1.8 | 31.3 | GO:0008038 | neuron recognition(GO:0008038) |
| 1.4 | 28.6 | GO:0001964 | startle response(GO:0001964) |
| 0.7 | 27.5 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 4.5 | 26.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 3.3 | 26.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 2.9 | 26.3 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 98.4 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.5 | 70.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 2.1 | 58.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 1.0 | 53.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.4 | 40.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 1.9 | 34.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.5 | 32.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 31.2 | GO:0045202 | synapse(GO:0045202) |
| 1.1 | 26.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.7 | 23.7 | GO:0043198 | dendritic shaft(GO:0043198) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 51.7 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 50.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 1.8 | 34.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 1.1 | 32.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 1.3 | 31.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.8 | 31.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 27.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.8 | 24.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 1.0 | 24.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 3.8 | 22.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 87.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 1.5 | 47.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 1.1 | 26.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.6 | 19.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 14.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.8 | 12.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.7 | 12.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 1.1 | 10.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 10.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.5 | 10.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 49.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 1.6 | 33.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 30.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 1.3 | 29.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 2.3 | 28.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 27.0 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.9 | 25.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 1.0 | 24.0 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 1.6 | 22.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.5 | 21.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |