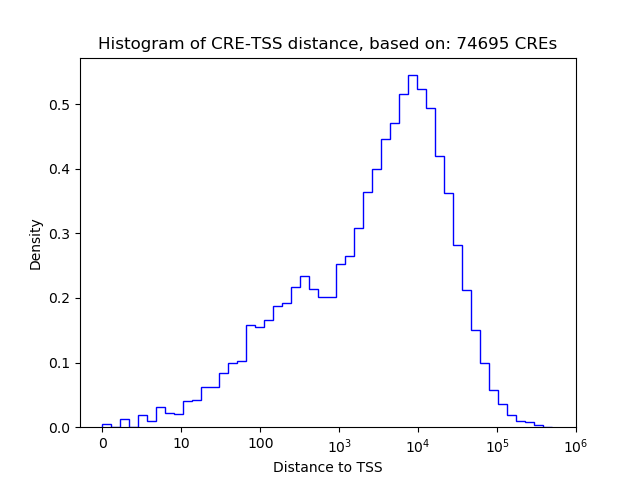
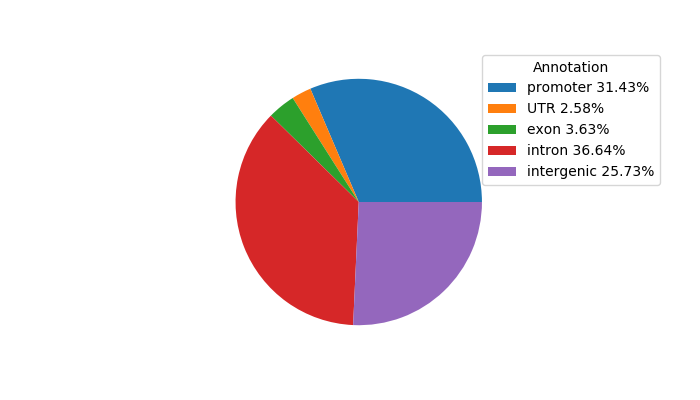
Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
Results for Hnf4g
Z-value: 9.33
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Hnf4g | chr3_3388435_3388601 | 119512 | 0.057180 | -0.68 | 9.9e-09 | Click! |
| Hnf4g | chr3_3526060_3526383 | 18191 | 0.191071 | 0.38 | 3.7e-03 | Click! |
| Hnf4g | chr3_3613607_3613959 | 20367 | 0.224067 | 0.35 | 9.7e-03 | Click! |
| Hnf4g | chr3_3507899_3508101 | 30 | 0.982958 | 0.34 | 1.1e-02 | Click! |
| Hnf4g | chr3_3645756_3645933 | 11694 | 0.265367 | 0.31 | 2.3e-02 | Click! |
Showing 1 to 5 of 5 entries
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_109557850_109558797 | 44.26 |
Crlf2 |
cytokine receptor-like factor 2 |
613 |
0.67 |
| chr15_79690079_79691459 | 30.51 |
Gtpbp1 |
GTP binding protein 1 |
76 |
0.92 |
| chr12_103863072_103863984 | 30.39 |
Serpina1a |
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
23 |
0.95 |
| chr18_3117365_3118622 | 29.39 |
Vmn1r238 |
vomeronasal 1 receptor, 238 |
5419 |
0.17 |
| chr18_79257106_79257504 | 29.19 |
Gm2116 |
predicted gene 2116 |
47857 |
0.15 |
| chr5_137530580_137532081 | 28.40 |
Gnb2 |
guanine nucleotide binding protein (G protein), beta 2 |
33 |
0.9 |
| chr10_127508848_127510720 | 28.16 |
Stac3 |
SH3 and cysteine rich domain 3 |
2559 |
0.15 |
| chr17_47923769_47925323 | 26.15 |
Foxp4 |
forkhead box P4 |
70 |
0.96 |
| chr17_34898151_34899707 | 25.91 |
Ehmt2 |
euchromatic histone lysine N-methyltransferase 2 |
25 |
0.87 |
| chr5_137786077_137787112 | 25.50 |
Mepce |
methylphosphate capping enzyme |
69 |
0.92 |
Network of associatons between targets according to the STRING database.
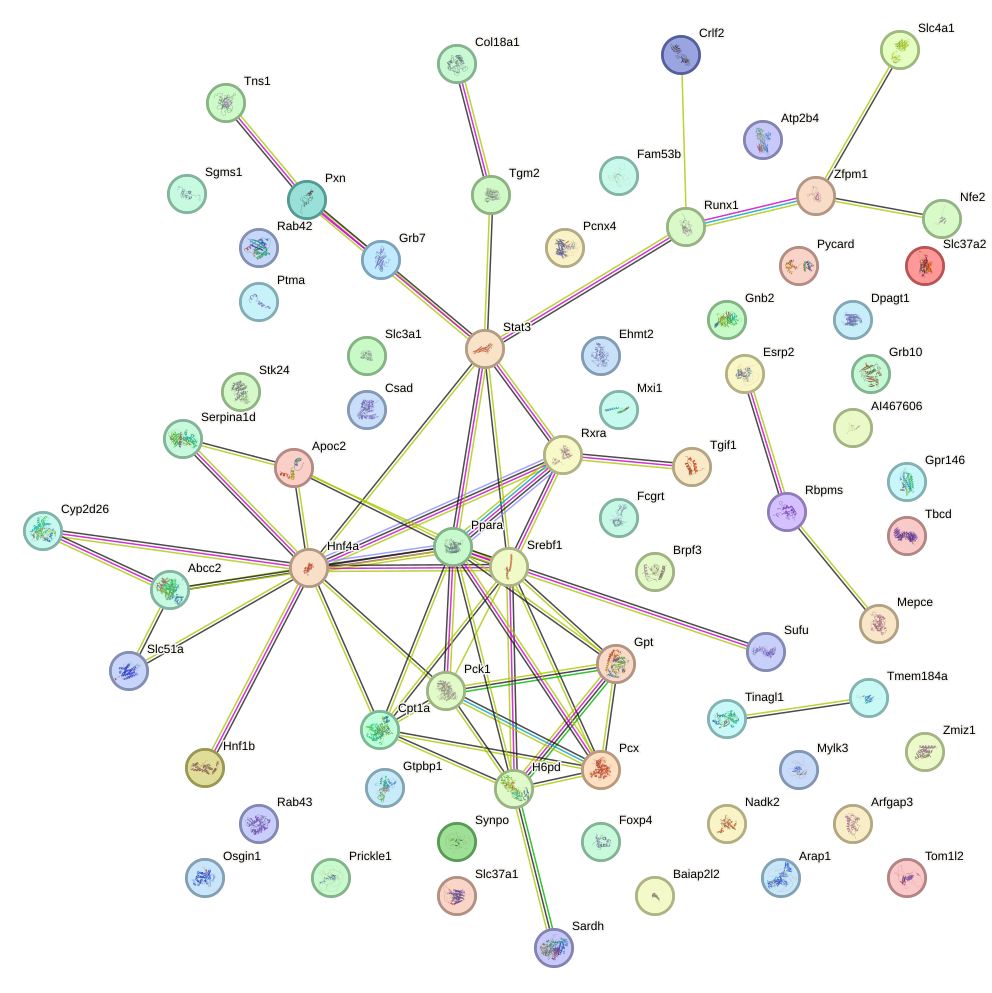
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 139.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.8 | 83.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 3.6 | 78.8 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 6.2 | 74.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 3.2 | 73.1 | GO:0014823 | response to activity(GO:0014823) |
| 5.0 | 69.7 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 1.1 | 59.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 3.9 | 59.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 4.8 | 57.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 4.5 | 54.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 1122.4 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.6 | 624.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.8 | 578.0 | GO:0005829 | cytosol(GO:0005829) |
| 1.8 | 454.2 | GO:0005924 | cell-substrate adherens junction(GO:0005924) |
| 0.4 | 265.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.8 | 251.2 | GO:0005730 | nucleolus(GO:0005730) |
| 1.4 | 215.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 2.4 | 210.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 2.0 | 188.3 | GO:0005903 | brush border(GO:0005903) |
| 2.6 | 155.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 267.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.5 | 213.3 | GO:0044822 | poly(A) RNA binding(GO:0044822) |
| 2.5 | 209.6 | GO:0002020 | protease binding(GO:0002020) |
| 1.0 | 164.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.8 | 113.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.7 | 102.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 1.4 | 89.6 | GO:0005178 | integrin binding(GO:0005178) |
| 1.0 | 87.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.9 | 85.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 2.0 | 83.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 206.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 5.0 | 130.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 2.5 | 126.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.6 | 116.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 3.4 | 110.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 2.5 | 109.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 2.7 | 102.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 3.2 | 100.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 3.8 | 98.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 3.6 | 93.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 184.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 3.9 | 113.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 1.3 | 113.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 1.2 | 111.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 1.9 | 110.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 3.5 | 103.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 4.5 | 103.6 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 6.3 | 101.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 2.4 | 96.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 2.1 | 93.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |